We will be happy to answer your questions.
PhenomeOne’s main system is designed for managing the entire plant research information of the organization. PhenomeOne is the perfect platform for users’ collaboration, making it their core organizational system for all research data, which can be updated on shared folders in a dedicated corporate directory tree, within the system. The, system covers all procedures, starting from pre-breeding, breeding, and variety testing, where each group has its own working space. For example, in this module…
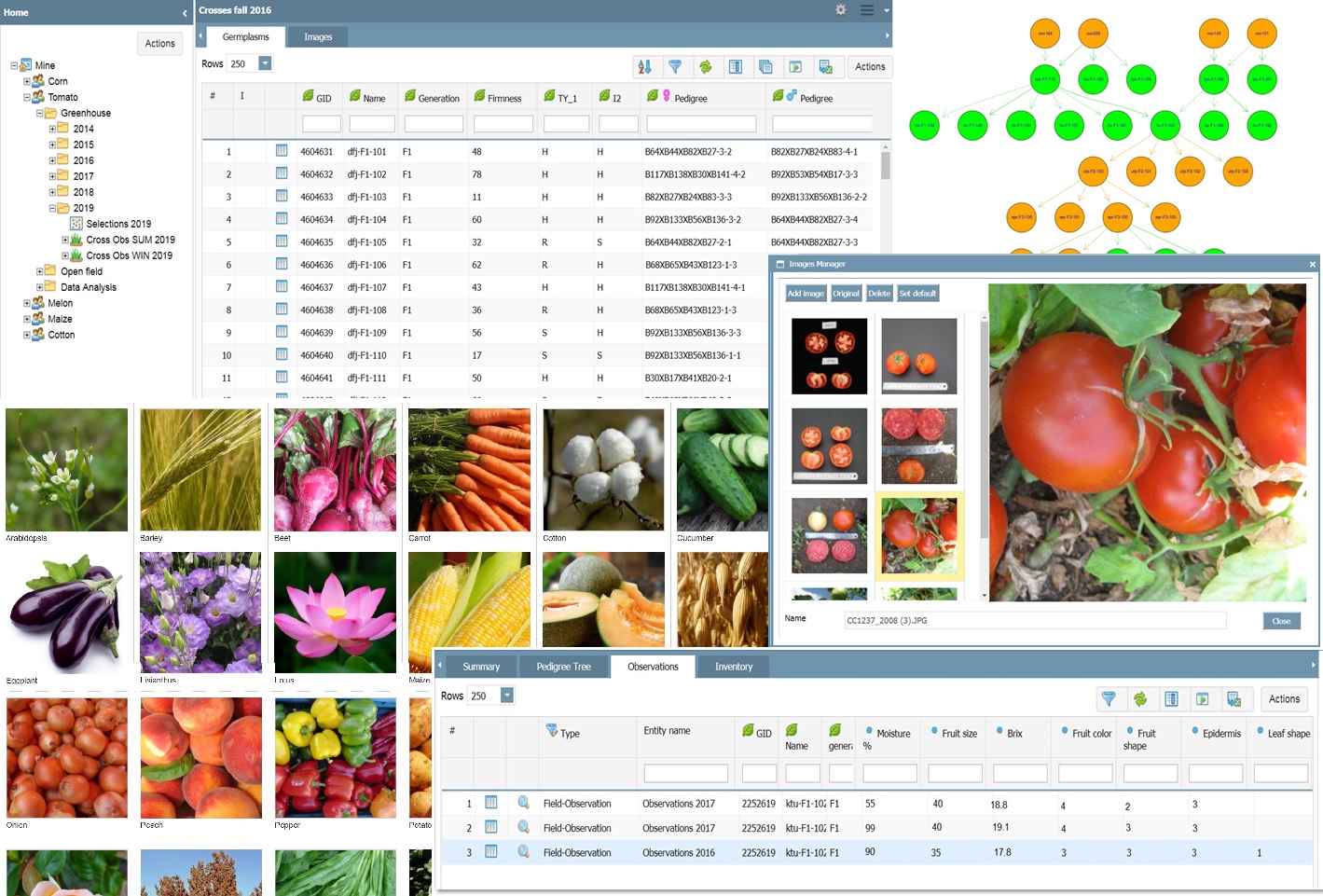
Introducing “Insights” – Phenome’s State-of-the-Art Analysis Module
We’re thrilled to introduce “Insights,” our advanced analysis and data visualization module, designed to extract valuable knowledge from data collected through our mobile app, PhenoTop, and our core software, PhenomOne. This module is specifically designed to compare germplasm (varieties) across a spectrum of environments or within replicated trials. The user can easily filter and compare between various programs, years, and locations. Results are displayed as interactive charts, supporting zoom-in and out, filtering and sorting, removing outliers, calculating significant differences, with an immediate response to user action.
PhenoTop is PhenomeOne’s new mobile application for smartphones and tablets (supporting iOS and Android devices). It is designed for offline and online data collection in fields, such as observations and selections, including images.
Fields are designed in the main system and then shared with relevant information to the mobile device. Thus, data can be collected offline and online and automatically and constantly synchronized when connected to the internet. The synchronization status is displayed for every field…
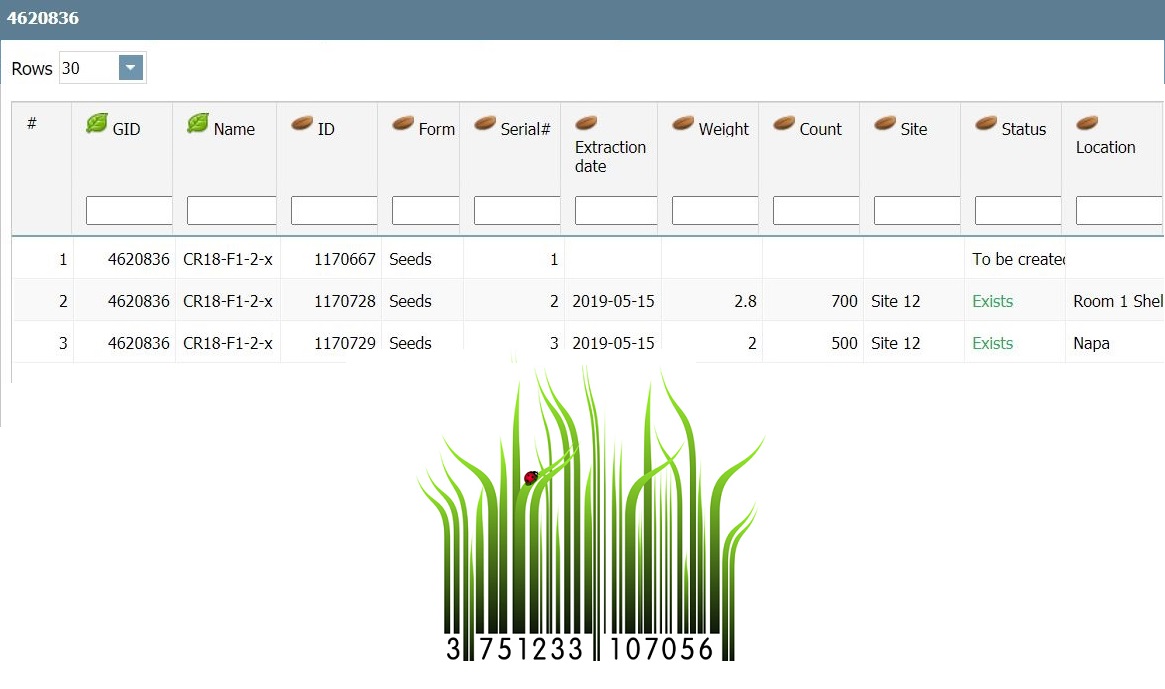
This module implements workflows integrating inventory activities of tracking and tracing Germplasms Lots throughout the product life cycle during research phases. Thus, inventory management becomes a part of sowing, harvesting, registering the harvested seeds and plants, updating quantities, possibly using barcodes or QR codes, electronic weights, and refractometers.
Germplasm in PhenomeOne refers to each unique genetic material, while Germplasm Lot refers to the physical entity of the genetics, typically seeds or plants. The same Germplasm (genetic entity) can have multiple Lots (physical seed packets), and each Lot can be stored in each location…
The analysis module presents quantitative and qualitative trait distributions, multivariate analysis, clustering, heat-maps, pedigree overlays with multiple phenotypes (or markers), combining ability, genotype by the environment, idiosyncratic heritability, statistical tests (T-test, F-test, analysis of variance), correlations, and more. Based on the R project for statistical computing, the analytics provide a rich repertoire of easily invoked tools.
Query results can be saved and accessed later, promoting team collaborations…
Phenome Networks, in collaboration with KeyGene, introduces PhenoGene, a game-changing solution for plant breeders to optimize breeding strategies.
The Module Purpose
The PhenoGene module provides the optimal breeding schedule that results in a specific ideal genotype (or ‘Ideotype’): one variety or a parent line that includes only desirable genetic markers obtained from several source parental lines.
PhenoGene calculates the optimal combination of alleles from up to 30 markers in several selected parent lines. Consequently, the module provides an optimal breeding plan for developing this one desired ideal genotype in the shortest, most cost-effective way. This optimal schedule will include detailed instructions to the crosses, selfs (selections), and the number of plants required, given several parents, markers, and other logistic and practical constraints.